Debugging and performances¶
Import and set-up
Import the library and toy data
[2]:
import pyxpcm
from pyxpcm.models import pcm
# Load a dataset to work with:
ds = pyxpcm.tutorial.open_dataset('argo').load()
# Define vertical axis and features to use:
z = np.arange(0.,-1000.,-10.)
features_pcm = {'temperature': z, 'salinity': z}
features_in_ds = {'temperature': 'TEMP', 'salinity': 'PSAL'}
Debugging¶
Use option debug to print log messages
[3]:
# Instantiate a new PCM:
m = pcm(K=8, features=features_pcm, debug=True)
# Fit with log:
m.fit(ds, features=features_in_ds);
> Start preprocessing for action 'fit'
> Preprocessing xarray dataset 'TEMP' as PCM feature 'temperature'
[<class 'xarray.core.dataarray.DataArray'>, <class 'dask.array.core.Array'>, ((7560,), (282,))] X RAVELED with success
Output axis is in the input axis, not need to interpolate, simple intersection
[<class 'xarray.core.dataarray.DataArray'>, <class 'dask.array.core.Array'>, ((7560,), (100,))] X INTERPOLATED with success)
[<class 'xarray.core.dataarray.DataArray'>, <class 'numpy.ndarray'>, None] X SCALED with success)
[<class 'xarray.core.dataarray.DataArray'>, <class 'numpy.ndarray'>, None] X REDUCED with success)
temperature pre-processed with success, [<class 'xarray.core.dataarray.DataArray'>, <class 'numpy.ndarray'>, None]
Homogenisation for fit of temperature
> Preprocessing xarray dataset 'PSAL' as PCM feature 'salinity'
[<class 'xarray.core.dataarray.DataArray'>, <class 'dask.array.core.Array'>, ((7560,), (282,))] X RAVELED with success
Output axis is in the input axis, not need to interpolate, simple intersection
[<class 'xarray.core.dataarray.DataArray'>, <class 'dask.array.core.Array'>, ((7560,), (100,))] X INTERPOLATED with success)
[<class 'xarray.core.dataarray.DataArray'>, <class 'numpy.ndarray'>, None] X SCALED with success)
[<class 'xarray.core.dataarray.DataArray'>, <class 'numpy.ndarray'>, None] X REDUCED with success)
salinity pre-processed with success, [<class 'xarray.core.dataarray.DataArray'>, <class 'numpy.ndarray'>, None]
Homogenisation for fit of salinity
Features array shape and type for xarray: (7560, 30) <class 'numpy.ndarray'> <class 'memoryview'>
> Preprocessing done, working with final X (<class 'xarray.core.dataarray.DataArray'>) array of shape: (7560, 30) and sampling dimensions: ['N_PROF']
Performance / Optimisation¶
Use timeit and timeit_verb to compute computation time of PCM operations
Times are accessible as a pandas Dataframe in timeit pyXpcm instance property.
The pyXpcm m.plot.timeit() plot method allows for a simple visualisation of times.
Time readings during execution¶
[4]:
# Create a PCM and execute methods:
m = pcm(K=8, features=features_pcm, timeit=True, timeit_verb=1)
m.fit(ds, features=features_in_ds);
fit.1-preprocess.1-mask: 20 ms
fit.1-preprocess.2-feature_temperature.1-ravel: 29 ms
fit.1-preprocess.2-feature_temperature.2-interp: 0 ms
fit.1-preprocess.2-feature_temperature.3-scale_fit: 7 ms
fit.1-preprocess.2-feature_temperature.4-scale_transform: 4 ms
fit.1-preprocess.2-feature_temperature.5-reduce_fit: 10 ms
fit.1-preprocess.2-feature_temperature.6-reduce_transform: 2 ms
fit.1-preprocess.2-feature_temperature.total: 55 ms
fit.1-preprocess: 56 ms
fit.1-preprocess.3-homogeniser: 1 ms
fit.1-preprocess.2-feature_salinity.1-ravel: 25 ms
fit.1-preprocess.2-feature_salinity.2-interp: 0 ms
fit.1-preprocess.2-feature_salinity.3-scale_fit: 7 ms
fit.1-preprocess.2-feature_salinity.4-scale_transform: 4 ms
fit.1-preprocess.2-feature_salinity.5-reduce_fit: 9 ms
fit.1-preprocess.2-feature_salinity.6-reduce_transform: 2 ms
fit.1-preprocess.2-feature_salinity.total: 51 ms
fit.1-preprocess: 51 ms
fit.1-preprocess.3-homogeniser: 1 ms
fit.1-preprocess.4-xarray: 0 ms
fit.1-preprocess: 132 ms
fit.fit: 2675 ms
fit.score: 9 ms
fit: 2817 ms
A posteriori Execution time analysis¶
[5]:
# Create a PCM and execute methods:
m = pcm(K=8, features=features_pcm, timeit=True, timeit_verb=0)
m.fit(ds, features=features_in_ds);
m.predict(ds, features=features_in_ds);
m.fit_predict(ds, features=features_in_ds);
Execution times are accessible through a dataframe with the pyxpcm.pcm.timeit property
[6]:
m.timeit
[6]:
Method Sub-method Sub-sub-method Sub-sub-sub-method
fit 1-preprocess 1-mask total 19.667864
2-feature_temperature 1-ravel 28.775930
2-interp 0.649929
3-scale_fit 10.529041
4-scale_transform 4.294872
5-reduce_fit 11.734009
6-reduce_transform 2.493382
total 58.599949
total 232.898712
3-homogeniser total 2.095938
2-feature_salinity 1-ravel 20.290136
2-interp 0.611067
3-scale_fit 7.843971
4-scale_transform 4.334688
5-reduce_fit 10.154247
6-reduce_transform 2.441883
total 45.794964
4-xarray total 0.997305
fit total 1721.768379
score total 8.548021
total 1859.453201
predict 1-preprocess 1-mask total 18.440008
2-feature_temperature 1-ravel 29.564142
2-interp 0.645876
3-scale_fit 0.000954
4-scale_transform 4.281998
5-reduce_fit 0.002146
6-reduce_transform 2.423048
total 37.017107
total 159.928322
...
2-feature_salinity 6-reduce_transform 2.339125
total 32.124043
4-xarray total 1.005888
predict total 8.524895
score total 8.650064
xarray total 6.422997
total 114.479065
fit_predict 1-preprocess 1-mask total 21.162987
2-feature_temperature 1-ravel 20.349026
2-interp 0.633001
3-scale_fit 0.001907
4-scale_transform 6.472826
5-reduce_fit 0.000954
6-reduce_transform 3.720999
total 31.296015
total 149.966955
3-homogeniser total 2.356052
2-feature_salinity 1-ravel 23.428917
2-interp 0.654697
3-scale_fit 0.000954
4-scale_transform 4.308939
5-reduce_fit 0.001192
6-reduce_transform 2.245188
total 30.743122
4-xarray total 0.961065
fit total 1801.964045
score total 8.421898
predict total 7.369041
xarray total 5.813122
total 1911.516190
Length: 66, dtype: float64
Visualisation help¶
To facilitate your analysis of execution times, you can use pyxpcm.plot.timeit().
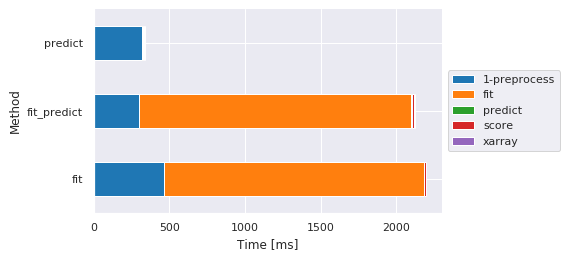
Main steps by method¶
[7]:
fig, ax, df = m.plot.timeit(group='Method', split='Sub-method', style='darkgrid') # Default group/split
df
[7]:
| Sub-method | 1-preprocess | fit | predict | score | xarray |
|---|---|---|---|---|---|
| Method | |||||
| fit | 464.207888 | 1721.768379 | NaN | 8.548021 | NaN |
| fit_predict | 298.304796 | 1801.964045 | 7.369041 | 8.421898 | 5.813122 |
| predict | 318.270445 | NaN | 8.524895 | 8.650064 | 6.422997 |
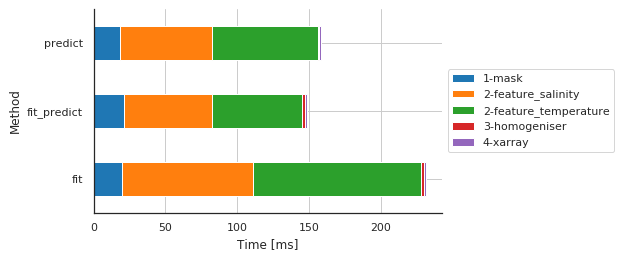
Preprocessing main steps by method¶
[8]:
fig, ax, df = m.plot.timeit(group='Method', split='Sub-sub-method')
df
[8]:
| Sub-sub-method | 1-mask | 2-feature_salinity | 2-feature_temperature | 3-homogeniser | 4-xarray |
|---|---|---|---|---|---|
| Method | |||||
| fit | 19.667864 | 91.470957 | 117.077112 | 2.095938 | 0.997305 |
| fit_predict | 21.162987 | 61.383009 | 62.474728 | 2.356052 | 0.961065 |
| predict | 18.440008 | 64.152002 | 73.935270 | 0.808954 | 1.005888 |
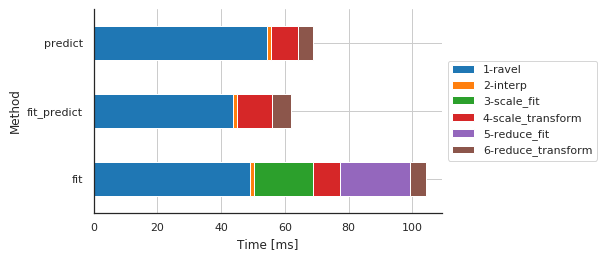
Preprocessing details by method¶
[9]:
fig, ax, df = m.plot.timeit(group='Method', split='Sub-sub-sub-method')
df
[9]:
| Sub-sub-sub-method | 1-ravel | 2-interp | 3-scale_fit | 4-scale_transform | 5-reduce_fit | 6-reduce_transform |
|---|---|---|---|---|---|---|
| Method | ||||||
| fit | 49.066067 | 1.260996 | 18.373013 | 8.629560 | 21.888256 | 4.935265 |
| fit_predict | 43.777943 | 1.287699 | 0.002861 | 10.781765 | 0.002146 | 5.966187 |
| predict | 54.390907 | 1.282930 | 0.002146 | 8.505106 | 0.002861 | 4.762173 |
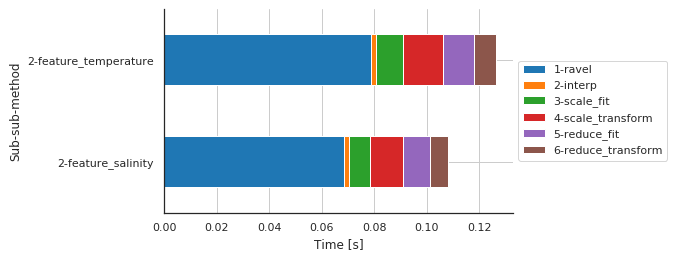
Preprocessing details by features¶
[10]:
fig, ax, df = m.plot.timeit(split='Sub-sub-sub-method', group='Sub-sub-method', unit='s')
df
[10]:
| Sub-sub-sub-method | 1-ravel | 2-interp | 3-scale_fit | 4-scale_transform | 5-reduce_fit | 6-reduce_transform |
|---|---|---|---|---|---|---|
| Sub-sub-method | ||||||
| 2-feature_salinity | 0.068546 | 0.001903 | 0.007846 | 0.012867 | 0.010156 | 0.007026 |
| 2-feature_temperature | 0.078689 | 0.001929 | 0.010532 | 0.015050 | 0.011737 | 0.008637 |