Debugging and performances¶
Import and set-up
Import the library and toy data
[2]:
import pyxpcm
from pyxpcm.models import pcm
# Load a dataset to work with:
ds = pyxpcm.tutorial.open_dataset('argo').load()
# Define vertical axis and features to use:
z = np.arange(0.,-1000.,-10.)
features_pcm = {'temperature': z, 'salinity': z}
features_in_ds = {'temperature': 'TEMP', 'salinity': 'PSAL'}
Debugging¶
Use option debug to print log messages
[3]:
# Instantiate a new PCM:
m = pcm(K=8, features=features_pcm, debug=True)
# Fit with log:
m.fit(ds, features=features_in_ds);
> Start preprocessing for action 'fit'
> Preprocessing xarray dataset 'TEMP' as PCM feature 'temperature'
[<class 'xarray.core.dataarray.DataArray'>, <class 'dask.array.core.Array'>, ((7560,), (282,))] X RAVELED with success
Output axis is in the input axis, not need to interpolate, simple intersection
[<class 'xarray.core.dataarray.DataArray'>, <class 'dask.array.core.Array'>, ((7560,), (100,))] X INTERPOLATED with success)
[<class 'xarray.core.dataarray.DataArray'>, <class 'numpy.ndarray'>, None] X SCALED with success)
[<class 'xarray.core.dataarray.DataArray'>, <class 'numpy.ndarray'>, None] X REDUCED with success)
temperature pre-processed with success, [<class 'xarray.core.dataarray.DataArray'>, <class 'numpy.ndarray'>, None]
Homogenisation for fit of temperature
> Preprocessing xarray dataset 'PSAL' as PCM feature 'salinity'
[<class 'xarray.core.dataarray.DataArray'>, <class 'dask.array.core.Array'>, ((7560,), (282,))] X RAVELED with success
Output axis is in the input axis, not need to interpolate, simple intersection
[<class 'xarray.core.dataarray.DataArray'>, <class 'dask.array.core.Array'>, ((7560,), (100,))] X INTERPOLATED with success)
[<class 'xarray.core.dataarray.DataArray'>, <class 'numpy.ndarray'>, None] X SCALED with success)
[<class 'xarray.core.dataarray.DataArray'>, <class 'numpy.ndarray'>, None] X REDUCED with success)
salinity pre-processed with success, [<class 'xarray.core.dataarray.DataArray'>, <class 'numpy.ndarray'>, None]
Homogenisation for fit of salinity
Features array shape and type for xarray: (7560, 30) <class 'numpy.ndarray'> <class 'memoryview'>
> Preprocessing done, working with final X (<class 'xarray.core.dataarray.DataArray'>) array of shape: (7560, 30) and sampling dimensions: ['N_PROF']
Performance / Optimisation¶
Use timeit and timeit_verb to compute computation time of PCM operations
Times are accessible as a pandas Dataframe in timeit pyXpcm instance property.
The pyXpcm m.plot.timeit() plot method allows for a simple visualisation of times.
Time readings during execution¶
[4]:
# Create a PCM and execute methods:
m = pcm(K=8, features=features_pcm, timeit=True, timeit_verb=1)
m.fit(ds, features=features_in_ds);
fit.1-preprocess.1-mask: 62 ms
fit.1-preprocess.2-feature_temperature.1-ravel: 27 ms
fit.1-preprocess.2-feature_temperature.2-interp: 2 ms
fit.1-preprocess.2-feature_temperature.3-scale_fit: 15 ms
fit.1-preprocess.2-feature_temperature.4-scale_transform: 6 ms
fit.1-preprocess.2-feature_temperature.5-reduce_fit: 21 ms
fit.1-preprocess.2-feature_temperature.6-reduce_transform: 7 ms
fit.1-preprocess.2-feature_temperature.total: 80 ms
fit.1-preprocess: 80 ms
fit.1-preprocess.3-homogeniser: 5 ms
fit.1-preprocess.2-feature_salinity.1-ravel: 32 ms
fit.1-preprocess.2-feature_salinity.2-interp: 1 ms
fit.1-preprocess.2-feature_salinity.3-scale_fit: 11 ms
fit.1-preprocess.2-feature_salinity.4-scale_transform: 5 ms
fit.1-preprocess.2-feature_salinity.5-reduce_fit: 18 ms
fit.1-preprocess.2-feature_salinity.6-reduce_transform: 4 ms
fit.1-preprocess.2-feature_salinity.total: 75 ms
fit.1-preprocess: 75 ms
fit.1-preprocess.3-homogeniser: 1 ms
fit.1-preprocess.4-xarray: 1 ms
fit.1-preprocess: 228 ms
fit.fit: 3400 ms
fit.score: 12 ms
fit: 3642 ms
A posteriori Execution time analysis¶
[5]:
# Create a PCM and execute methods:
m = pcm(K=8, features=features_pcm, timeit=True, timeit_verb=0)
m.fit(ds, features=features_in_ds);
m.predict(ds, features=features_in_ds);
m.fit_predict(ds, features=features_in_ds);
Execution times are accessible through a dataframe with the pyxpcm.pcm.timeit property
[6]:
m.timeit
[6]:
Method Sub-method Sub-sub-method Sub-sub-sub-method
fit 1-preprocess 1-mask total 60.623884
2-feature_temperature 1-ravel 29.070854
2-interp 1.361847
3-scale_fit 24.303198
4-scale_transform 5.542994
5-reduce_fit 17.215014
6-reduce_transform 4.530907
total 82.225800
total 405.465841
3-homogeniser total 3.330231
2-feature_salinity 1-ravel 33.647060
2-interp 1.427889
3-scale_fit 19.104004
4-scale_transform 16.283989
5-reduce_fit 13.432264
6-reduce_transform 3.180981
total 87.301970
4-xarray total 1.182079
fit total 1668.042660
score total 14.346838
total 1918.222189
predict 1-preprocess 1-mask total 64.723015
2-feature_temperature 1-ravel 28.513908
2-interp 1.239061
3-scale_fit 0.003099
4-scale_transform 7.060051
5-reduce_fit 0.002146
6-reduce_transform 2.730846
total 39.700031
total 235.766172
...
2-feature_salinity 6-reduce_transform 2.788067
total 44.227123
4-xarray total 1.113892
predict total 10.058880
score total 11.398077
xarray total 11.323929
total 184.562922
fit_predict 1-preprocess 1-mask total 64.216852
2-feature_temperature 1-ravel 26.321888
2-interp 1.183033
3-scale_fit 0.001907
4-scale_transform 5.228996
5-reduce_fit 0.000954
6-reduce_transform 2.723217
total 35.592079
total 224.620104
3-homogeniser total 2.858639
2-feature_salinity 1-ravel 29.989958
2-interp 1.201153
3-scale_fit 0.000954
4-scale_transform 5.232811
5-reduce_fit 0.001907
6-reduce_transform 4.884958
total 41.451693
4-xarray total 1.657963
fit total 2717.261076
score total 11.775970
predict total 10.827065
xarray total 10.989189
total 2898.393869
Length: 66, dtype: float64
Visualisation help¶
To facilitate your analysis of execution times, you can use pyxpcm.plot.timeit().
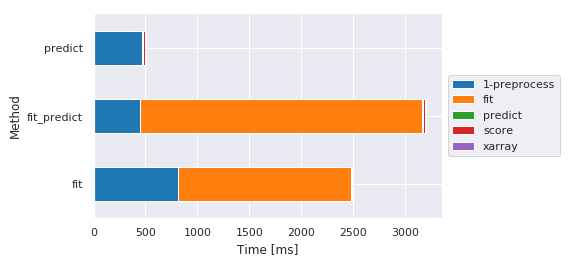
Main steps by method¶
[7]:
fig, ax, df = m.plot.timeit(group='Method', split='Sub-method', style='darkgrid') # Default group/split
df
[7]:
| Sub-method | 1-preprocess | fit | predict | score | xarray |
|---|---|---|---|---|---|
| Method | |||||
| fit | 809.230804 | 1668.042660 | NaN | 14.346838 | NaN |
| fit_predict | 447.169065 | 2717.261076 | 10.827065 | 11.775970 | 10.989189 |
| predict | 469.947577 | NaN | 10.058880 | 11.398077 | 11.323929 |
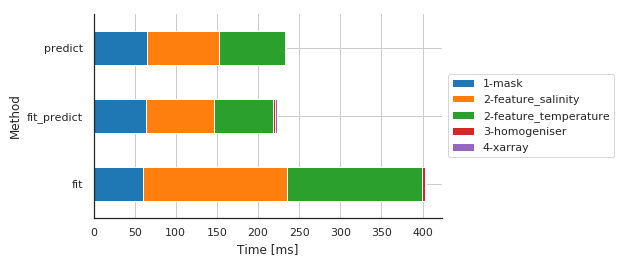
Preprocessing main steps by method¶
[8]:
fig, ax, df = m.plot.timeit(group='Method', split='Sub-sub-method')
df
[8]:
| Sub-sub-method | 1-mask | 2-feature_salinity | 2-feature_temperature | 3-homogeniser | 4-xarray |
|---|---|---|---|---|---|
| Method | |||||
| fit | 60.623884 | 174.378157 | 164.250612 | 3.330231 | 1.182079 |
| fit_predict | 64.216852 | 82.763433 | 71.052074 | 2.858639 | 1.657963 |
| predict | 64.723015 | 88.269234 | 79.249144 | 0.826120 | 1.113892 |
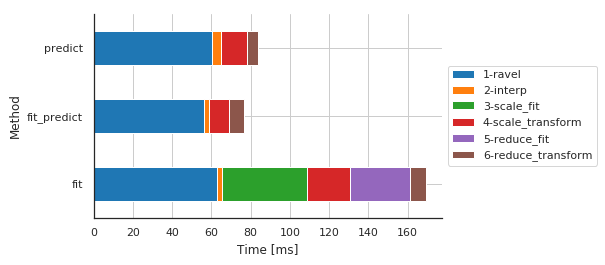
Preprocessing details by method¶
[9]:
fig, ax, df = m.plot.timeit(group='Method', split='Sub-sub-sub-method')
df
[9]:
| Sub-sub-sub-method | 1-ravel | 2-interp | 3-scale_fit | 4-scale_transform | 5-reduce_fit | 6-reduce_transform |
|---|---|---|---|---|---|---|
| Method | ||||||
| fit | 62.717915 | 2.789736 | 43.407202 | 21.826982 | 30.647278 | 7.711887 |
| fit_predict | 56.311846 | 2.384186 | 0.002861 | 10.461807 | 0.002861 | 7.608175 |
| predict | 60.415030 | 4.472017 | 0.005245 | 13.175964 | 0.004053 | 5.518913 |
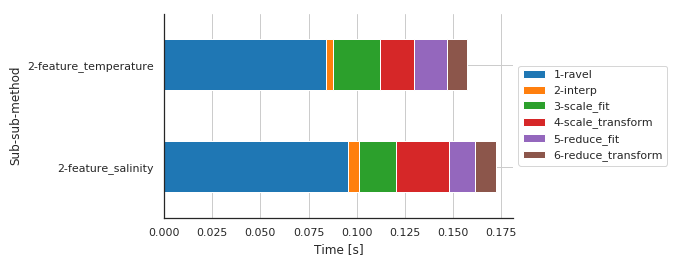
Preprocessing details by features¶
[10]:
fig, ax, df = m.plot.timeit(split='Sub-sub-sub-method', group='Sub-sub-method', unit='s')
df
[10]:
| Sub-sub-sub-method | 1-ravel | 2-interp | 3-scale_fit | 4-scale_transform | 5-reduce_fit | 6-reduce_transform |
|---|---|---|---|---|---|---|
| Sub-sub-method | ||||||
| 2-feature_salinity | 0.095538 | 0.005862 | 0.019107 | 0.027633 | 0.013436 | 0.010854 |
| 2-feature_temperature | 0.083907 | 0.003784 | 0.024308 | 0.017832 | 0.017218 | 0.009985 |