Debugging and performances
Import and set-up
Import the library and toy data
[2]:
import pyxpcm
from pyxpcm.models import pcm
# Load a dataset to work with:
ds = pyxpcm.tutorial.open_dataset('argo').load()
# Define vertical axis and features to use:
z = np.arange(0.,-1000.,-10.)
features_pcm = {'temperature': z, 'salinity': z}
features_in_ds = {'temperature': 'TEMP', 'salinity': 'PSAL'}
Debugging
Use option debug to print log messages
[3]:
# Instantiate a new PCM:
m = pcm(K=8, features=features_pcm, debug=True)
# Fit with log:
m.fit(ds, features=features_in_ds);
> Start preprocessing for action 'fit'
> Preprocessing xarray dataset 'TEMP' as PCM feature 'temperature'
X RAVELED with success [<class 'xarray.core.dataarray.DataArray'>, <class 'dask.array.core.Array'>, ((7560,), (282,))]
Output axis is in the input axis, not need to interpolate, simple intersection
X INTERPOLATED with success [<class 'xarray.core.dataarray.DataArray'>, <class 'dask.array.core.Array'>, ((7560,), (100,))]
X SCALED with success) [<class 'xarray.core.dataarray.DataArray'>, <class 'numpy.ndarray'>, None]
X REDUCED with success) [<class 'xarray.core.dataarray.DataArray'>, <class 'numpy.ndarray'>, None]
temperature pre-processed with success, [<class 'xarray.core.dataarray.DataArray'>, <class 'numpy.ndarray'>, None]
Homogenisation for fit of temperature
> Preprocessing xarray dataset 'PSAL' as PCM feature 'salinity'
X RAVELED with success [<class 'xarray.core.dataarray.DataArray'>, <class 'dask.array.core.Array'>, ((7560,), (282,))]
Output axis is in the input axis, not need to interpolate, simple intersection
X INTERPOLATED with success [<class 'xarray.core.dataarray.DataArray'>, <class 'dask.array.core.Array'>, ((7560,), (100,))]
X SCALED with success) [<class 'xarray.core.dataarray.DataArray'>, <class 'numpy.ndarray'>, None]
X REDUCED with success) [<class 'xarray.core.dataarray.DataArray'>, <class 'numpy.ndarray'>, None]
salinity pre-processed with success, [<class 'xarray.core.dataarray.DataArray'>, <class 'numpy.ndarray'>, None]
Homogenisation for fit of salinity
Features array shape and type for xarray: (7560, 30) <class 'numpy.ndarray'> <class 'memoryview'>
> Preprocessing done, working with final X (<class 'xarray.core.dataarray.DataArray'>) array of shape: (7560, 30) and sampling dimensions: ['N_PROF']
Performance / Optimisation
Use timeit and timeit_verb to compute computation time of PCM operations
Times are accessible as a pandas Dataframe in timeit pyXpcm instance property.
The pyXpcm m.plot.timeit() plot method allows for a simple visualisation of times.
Time readings during execution
[4]:
# Create a PCM and execute methods:
m = pcm(K=8, features=features_pcm, timeit=True, timeit_verb=1)
m.fit(ds, features=features_in_ds);
fit.1-preprocess.1-mask: 26 ms
fit.1-preprocess.2-feature_temperature.1-ravel: 48 ms
fit.1-preprocess.2-feature_temperature.2-interp: 2 ms
fit.1-preprocess.2-feature_temperature.3-scale_fit: 14 ms
fit.1-preprocess.2-feature_temperature.4-scale_transform: 8 ms
fit.1-preprocess.2-feature_temperature.5-reduce_fit: 22 ms
fit.1-preprocess.2-feature_temperature.6-reduce_transform: 5 ms
fit.1-preprocess.2-feature_temperature.total: 103 ms
fit.1-preprocess: 103 ms
fit.1-preprocess.3-homogeniser: 1 ms
fit.1-preprocess.2-feature_salinity.1-ravel: 42 ms
fit.1-preprocess.2-feature_salinity.2-interp: 1 ms
fit.1-preprocess.2-feature_salinity.3-scale_fit: 12 ms
fit.1-preprocess.2-feature_salinity.4-scale_transform: 10 ms
fit.1-preprocess.2-feature_salinity.5-reduce_fit: 14 ms
fit.1-preprocess.2-feature_salinity.6-reduce_transform: 3 ms
fit.1-preprocess.2-feature_salinity.total: 85 ms
fit.1-preprocess: 85 ms
fit.1-preprocess.3-homogeniser: 1 ms
fit.1-preprocess.4-xarray: 1 ms
fit.1-preprocess: 225 ms
fit.fit: 2206 ms
fit.score: 10 ms
fit: 2442 ms
A posteriori Execution time analysis
[5]:
# Create a PCM and execute methods:
m = pcm(K=8, features=features_pcm, timeit=True, timeit_verb=0)
m.fit(ds, features=features_in_ds);
m.predict(ds, features=features_in_ds);
m.fit_predict(ds, features=features_in_ds);
Execution times are accessible through a dataframe with the pyxpcm.pcm.timeit property
[6]:
m.timeit
[6]:
Method Sub-method Sub-sub-method Sub-sub-sub-method
fit 1-preprocess 1-mask total 19.836187
2-feature_temperature 1-ravel 32.550097
2-interp 0.828981
3-scale_fit 9.926319
4-scale_transform 5.232811
...
fit_predict fit total 737.503052
score total 8.855104
predict total 9.073734
xarray total 8.368969
total 882.753134
Length: 66, dtype: float64
Visualisation help
To facilitate your analysis of execution times, you can use pyxpcm.plot.timeit().
Main steps by method
[7]:
fig, ax, df = m.plot.timeit(group='Method', split='Sub-method', style='darkgrid') # Default group/split
df
[7]:
| Sub-method | 1-preprocess | fit | predict | score | xarray |
|---|---|---|---|---|---|
| Method | |||||
| fit | 556.826353 | 1123.903990 | NaN | 10.521889 | NaN |
| fit_predict | 416.832924 | 737.503052 | 9.073734 | 8.855104 | 8.368969 |
| predict | 402.269125 | NaN | 9.886980 | 9.831905 | 8.663177 |
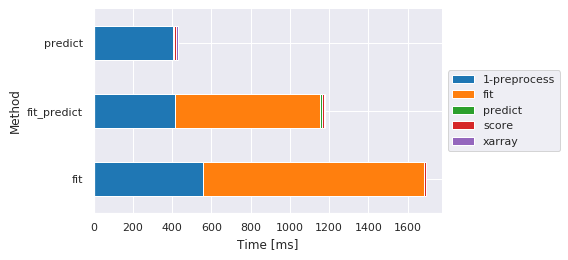
Preprocessing main steps by method
[8]:
fig, ax, df = m.plot.timeit(group='Method', split='Sub-sub-method')
df
[8]:
| Sub-sub-method | 1-mask | 2-feature_salinity | 2-feature_temperature | 3-homogeniser | 4-xarray |
|---|---|---|---|---|---|
| Method | |||||
| fit | 19.836187 | 122.781515 | 130.059242 | 2.494097 | 2.145052 |
| fit_predict | 20.085096 | 93.130827 | 89.278936 | 3.419876 | 1.243114 |
| predict | 24.972916 | 73.968887 | 98.288298 | 1.749992 | 1.240015 |
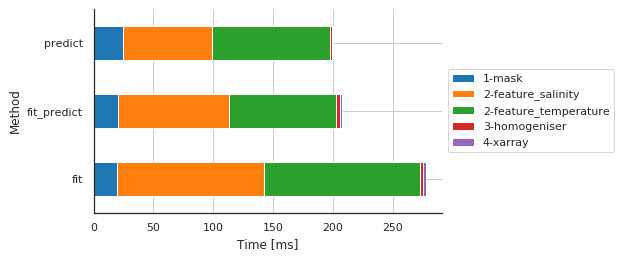
Preprocessing details by method
[9]:
fig, ax, df = m.plot.timeit(group='Method', split='Sub-sub-sub-method')
df
[9]:
| Sub-sub-sub-method | 1-ravel | 2-interp | 3-scale_fit | 4-scale_transform | 5-reduce_fit | 6-reduce_transform |
|---|---|---|---|---|---|---|
| Method | ||||||
| fit | 63.939333 | 1.705170 | 18.492460 | 10.242701 | 25.922775 | 5.970001 |
| fit_predict | 67.147017 | 1.942873 | 0.005245 | 12.181759 | 0.003815 | 9.781122 |
| predict | 67.429066 | 1.815319 | 0.004053 | 10.471821 | 0.003099 | 6.281853 |
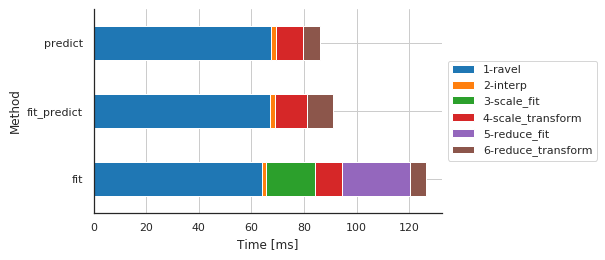
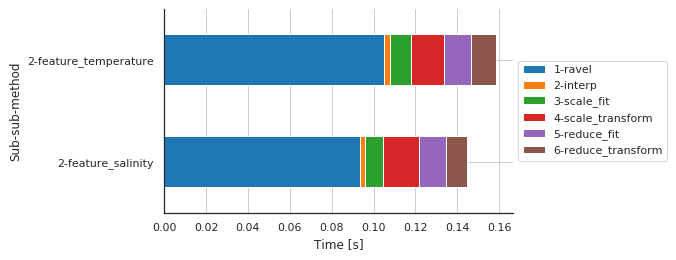
Preprocessing details by features
[10]:
fig, ax, df = m.plot.timeit(split='Sub-sub-sub-method', group='Sub-sub-method', unit='s')
df
[10]:
| Sub-sub-sub-method | 1-ravel | 2-interp | 3-scale_fit | 4-scale_transform | 5-reduce_fit | 6-reduce_transform |
|---|---|---|---|---|---|---|
| Sub-sub-method | ||||||
| 2-feature_salinity | 0.093312 | 0.002782 | 0.008571 | 0.017127 | 0.012878 | 0.010059 |
| 2-feature_temperature | 0.105203 | 0.002681 | 0.009930 | 0.015770 | 0.013052 | 0.011974 |